بازدید:37673 بروزرسانی: 20-12-1402
Click here or contact me to download PARS software.
PARS is designed for calculating the number of sequence fragments (mono, di and three-peptides), with assigning their conformation in secondary structure terms. As input, you need the structural file format, *.DSSP, for your structure. This file format is a plain text file format with DSSP extension, containing necessary secondary structure data for any residues.
Working with PDB structural file format - obtained from Protein Data Bank, you have to convert it to DSSP format. This can be done in some ways. See these pages for more information:
http://www.cmbi.ru.nl/dssp.html
or
A DSSP file format sample is here.
After preparing your input, you can use PARS software. Here is graphical step by step help:
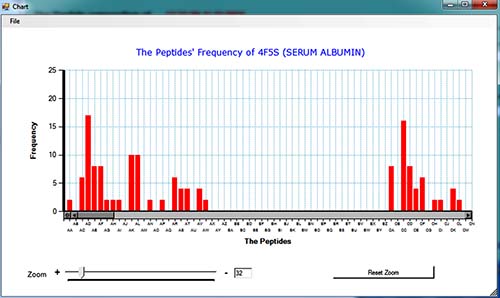
(Crystal Structure of Bovine Serum Albumin, PDB ID: 4f5s explained here)
1
-Load your input:
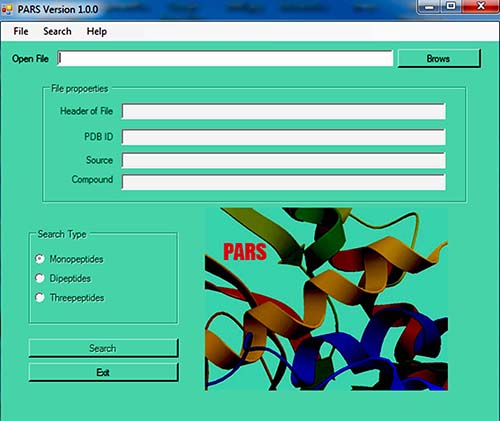
Calculate fragment frequencies and save the results:
A sample of graphical output:
Also, the outputs are savable as two format, both in PARS extensions. (Format1, Format2)
For any question, specially reporting bugs, contact: heshmati@znu.ac.ir or bioinf.comp.biol@gmail.com

